Rothbart
Laboratory
 Chromatin and Epigenetic Regulation
Chromatin and Epigenetic Regulation
The Rothbart Laboratory leverages in vitro and cellular biochemistry, computational and molecular biophysics, pharmacology, genomics and proteomics to uncover basic molecular and cellular mechanisms controlling chromatin accessibility, interaction and function. We are particularly interested in understanding how histone post-translational modifications and DNA methylation work together as a language or “code” that is read and interpreted by specialized proteins to orchestrate the dynamic functions associated with chromatin.
Importantly, large-scale genomic and epigenomic sequencing efforts, spearheaded in part by laboratories in VAI’s Department of Epigenetics, are revealing that deregulation of the cellular machineries responsible for regulating chromatin function contribute to the initiation and progression of multiple human diseases, including cancer. Unlike genetic abnormalities, DNA and histone modifications are reversible, making the writers, erasers and readers of these marks attractive targets for therapeutic intervention. Given this, our long-term goal is to translate the basic research performed in the lab toward novel chromatin and epigenetic target identification and drug discovery.
Our mission is to implement innovative scientific approaches and develop useful tools, reagents and therapeutics that advance our understanding of chromatin function in human health and disease. Central to the success of this mission is our dedication to the education and training of scientists to prepare them to become leaders in the scientific community.
About Epigenetics
Think of epigenetics as a set of control switches that operate in every cell in the body. These switches help pack and unpack DNA, the spiraling molecule that acts as the blueprint for life.
In order for DNA to fit into the cell, it must be tightly balled up much like a crumpled piece of paper. For the information contained within the genetic blueprint to be read, however, it must be unfolded. Epigenetic mechanisms help open up the DNA, allowing access to the packaged blueprints contained within.
Epigenetic mechanisms determine the identity and function of each cell during development; some become heart cells, some become brain cells, and so on. Epigenetics help keep these cell types in their determined states.
In recent years, it’s become clear that when epigenetic control switches go awry, it can lead to diseases such as cancer, Parkinson’s and others.
The goal of the work we do in the Rothbart Lab is to understand, at the basic mechanistic level, how these epigenetic control switches work so we can harness them to better treat disease.
While all cells in the human body share a virtually identical genome, layers of regulation outside the genetic information encoded in the DNA (called epigenetics) program cells and give them unique identity. A complete copy of the human genome is tightly packaged inside every cell nucleus by wrapping around histone proteins.
This high level of DNA compaction presents a potential problem, as the underlying sequence must remain accessible to the vast protein machineries that utilize it for critical biological functions such as DNA replication, transcription and repair—all of which must occur at the appropriate place and time to promote cell growth, differentiation and proper organismal development.
Recent News
Learn More
Fighting to outsmart cancer: A Q&A with graduate student Yanqing Liu
Using viral mimicry to help the immune system see — and fight — cancer

Pinpointing weaknesses in cancer’s armor
Chomiak AA, Tiedemann RL, Liu Y, Kong X, Cui Y, Wiseman AK, Thurlow KE, Cornett EM, Topper MJ, Baylin SB, Rothbart SB. 2024. Select EZH2 inhibitors enhance viral mimicry effects of DNMT inhibition through a mechanism involving NFAT:AP-1 signaling. Sci Adv 10(13).
Kong X*, Chen J*, Xie W* et al. 2019. Defining UHRF1 domains that support maintenance of human colon cancer DNA methylation and oncogenic properties. Cancer Cell.
Our Impact
We’re raising thousands to save millions.
We’re turning hope into action for the millions of people around the world affected by diseases like cancer and Parkinson’s. Find out how you can help us make a difference.
- 122 peer-reviewed papers published in 2024, 63 of which were in high-impact journals
- 15 VAI-SU2C Epigenetics Dream Team clinical trials launched to date
- 10 clinical trials co-funded by VAI & Cure Parkinson's (out of 41 total International Linked Clinical Trials Program trials)
Scott Rothbart, Ph.D.
Professor, Department of Epigenetics
Areas of Expertise
Chromatin biochemistry, cancer epigenetics, functional proteomics
Biography
Dr. Scott Rothbart is an internationally recognized expert in the field of epigenetics, particularly in the area of chromatin regulation through histone post-translational modifications. He earned a B.S. in food science and human nutrition from the University of Florida, followed by a Ph.D. in pharmacology and toxicology from Virginia Commonwealth University in the lab of Dr. Rick Moran. Dr. Rothbart then completed a postdoctoral fellowship in Dr. Brian Strahl’s lab at the University of North Carolina at Chapel Hill. He established his lab at Van Andel Institute in 2015 and rose through the ranks to full professor in 2023. Dr. Rothbart is also a basic science investigator on the Van Andel Institute–Stand Up To Cancer Epigenetics Dream Team and co-director of the VAI Cancer Epigenetics Training Program. Dr. Rothbart’s work has contributed fundamental insights into mechanisms of epigenetic regulation and has introduced new tools and methodologies to the field. He is the recipient of numerous awards and honors, including a K99/R00 career development award from the National Cancer Institute, an R35 Maximizing Investigators’ Research Award (MIRA) from the National Institute of General Medical Sciences, and a Research Scholar Award from the American Cancer Society.
We are an interdisciplinary team that studies the basic biochemical and biophysical processes that drive the regulation of chromatin structure. As part of this pursuit, we are developing new and innovative in vitro systems, “omic” technologies and computational approaches to answer several questions:
- How are the writers and erasers of chromatin modifications regulated?
- How do nuclear proteins and their complexes interface with (i.e., read) epigenetic marks to perform their chromatin regulatory functions?
- How does deregulation of chromatin signaling contribute to human pathologies, including cancer, cardiovascular disease and autoimmune disorders?
Mechanisms regulating the epigenetic inheritance of DNA methylation
DNA methylation in mammals occurs primarily at cytosine residues of CpG dinucleotides. The faithful mitotic inheritance of DNA methylation patterns is essential for mammalian development and has been well studied for its role in the stable repression of transposons and endogenous retroviruses, gene silencing, genomic imprinting and X-chromosome inactivation. DNA methylation represses transcription directly by inhibiting the retention of sequence-specific DNA-binding transcription factors and indirectly through the recruitment of co-repressor complexes. DNA methylation patterns are established by the de novo methyltransferases DNMT3A and DNMT3B and are epigenetically inherited through somatic cell divisions by the replication-coupled maintenance DNA methyltransferase DNMT1. DNA methylation is erased passively with cell division in the absence of DNMT activity and actively by the TET family of DNA hydroxylases.
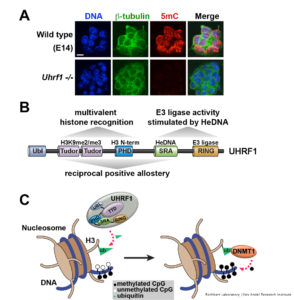
The protein UHRF1 has emerged in recent years as a key regulator of DNA methylation inheritance. Mice lacking either DNMT1 or UHRF1 show similar phenotypes of global DNA hypomethylation (Figure 1A) and early embryonic death. We and others have shown that the DNA methylation regulatory function of UHRF1 depends on its ability to bind chromatin (Rothbart et al 2012 Nat Struc Mol Biol; Rothbart et al 2013 Genes Dev), where it facilitates histone H3K18 and H3K23 mono-ubiquitination, a docking site for DNMT1 (Harrison et al 2016 eLife). Our studies show that the UHRF1 reader and writer domains function in a coordinated manner to engage the histone and DNA components of chromatin, to direct its ubiquitin ligase activity towards H3K18 and H3K23, and to regulate the epigenetic inheritance of DNA methylation (Vaughan et al 2018 Nuc Acids Res; Vaughan et al 2018 Proc Natl Acad Sci USA) (Figure 1B-C).
Notably, our findings assign a key biological function to hemi-methylated DNA as a member of an emerging class of non-protein ligand activators of E3 ligases, and they define the relationship between the enzymatic, histone and DNA-binding activities of UHRF1 as a multidomain epigenetic regulator of DNA methylation inheritance. Our studies present the first demonstration of an E3 ubiquitin ligase being activated by DNA binding and are the first to show that an epigenetic modification controls E3 ligase function. Using UHRF1 as a model system, we are working to define the underlying principles of multivalent engagement and allosteric regulation of enzymatic activity on chromatin, and we are applying these conceptual advances to the study of other biologically interesting and disease-relevant chromatin regulators.
The appreciation of UHRF1 as a key regulator of DNA methylation inheritance suggests UHRF1 antagonism as an innovative therapeutic strategy to target aberrant DNA methylation patterning, which is a hallmark of human cancers. The role of UHRF1 in tumorigenesis and the therapeutic potential of UHRF1 inhibition have now become major translational foci of the lab. This translational potential also drives our continuing basic studies of UHRF1 function and mechanism.
Development of proteomics tools for the study of histone and non-histone PTM regulators
Histone post-translational modifications (PTMs) and their combinations function in two ways:
- By directly altering the physical structure of chromatin
- As docking sites for reader domain-containing proteins that elicit selective effects on gene expression and other chromatin-templated processes.
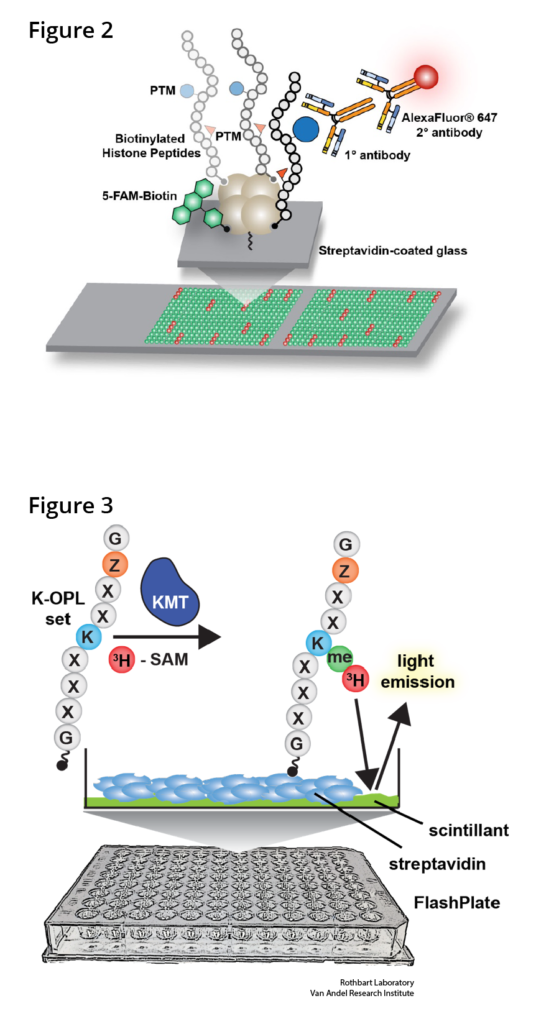
We contributed to the development of peptide microarray technology to query the activities of known and putative histone PTM regulators (Rothbart et al 2012 Methods Enzymol; Cornett et al 2016 Methods Enzymol) (Figure 2). We have used this platform extensively to characterize the reader, writer and eraser activities of these regulators and the behavior of antibodies that recognize histones and their PTMs (Rothbart et al 2015 Mol Cell; Shah et al 2018 Mol Cell). Because histone PTM antibodies are key reagents for immunoblotting, immunostaining and chromatin immunoprecipitation (ChIP) experiments, we continually profile the behavior of the most used and cited commercial histone PTM antibodies and compile this information into a public database (www.histoneantibodies.com). This interactive web resource is a valuable tool for the research community that routinely uses these antibodies in a wide range of applications.
From large-scale cancer exome sequencing studies, we now appreciate that chromatin regulators (including writers, erasers and readers of histone PTMs) are among the most frequently amplified and mutated genes in human cancers. This has sparked tremendous interest from the cancer and chromatin biology communities and has led to the development of small-molecule inhibitors that are in various phases of preclinical and clinical development. Defining the specificities of histone (and non-histone) PTM writers, erasers and readers is therefore critical for understanding the mechanisms of action of emerging epigenetic drugs, for rationalizing potential combination therapies and for identifying reliable biomarkers of clinical activity. To this end, we are developing new functional proteomics platforms that query the activities methyllysine writers (Cornett et al 2018 Science Adv) (Figure 3), erasers and readers. We consider these screening efforts analogous to genome and epigenome roadmap initiatives, where cartography and public data dissemination provide a foundation for later molecular studies. Datasets and bioinformatics tools generated from these studies are being used to facilitate comprehensive mapping of methylation sites throughout the human proteome, to promote the discovery of previously unknown substrates and interaction networks, and to identify optimal substrates for drug screening and structural studies.
Selected Publications
For comprehensive lists of Dr. Rothbart’s publications please visit his Google Scholar page or his PubMed.
2025
Akano I, Hebert JM, Tiedemann RL, Gao Q, Xiao Y, Prescott NA, Liu Y, Tan KS, Bastle RM, Ramakrishnan A, Maze I, Sidoli S, Koche RP, Ganesh K, Rothbart SB, David Y. 2025. The SWI/SNF-related protein SMARCA3 is a histone H3K23 ubiquitin ligase that regulates H3K9me3 in cancer. Mol Cell.
Bai W, Xu J, Gu W, Wang D, Cui Y, Rong W, Du X, Li X, Xia C, Gan Q, He G, Guo H, Deng J, Wu Y, Yen RWC, Yegnasubramanian S, Rothbart SB, Luo C, Wu L, Liu J, Baylin SB, Kong X. 2025. Defining ortholog-specific UHRF1 inhibition by STELLA for cancer therapy. Nat Commun 16:474.
2024
Liu Y, Hrit JA, Chomiak AA, Stransky S, Hoffman JR, Tiedemann RL, Wiseman AK, Kariapper LS, Dickson BM, Worden EJ, Fry CJ, Sidoli S, Rothbart SB. 2024. DNA hypomethylation promotes UHRF1-and SUV39H1/H2-dependent crosstalk between H3K18ub and H3K9me3 to reinforce heterochromatin states. Mol Cell.
Tiedemann RL, Hrit J, Du Q, Wiseman AK, Eden HE, Dickson BM, Kong X, Chomiak AA, Vaughan RM, Tibben BM, Hebert JM, David Y, Zhou W, Baylin SB, Jones PA, Clark SJ, Rothbart SB. 2024. UHRF1 ubiquitin ligase activity supports the maintenance of low-density CpG methylation. Nucleic Acids Res:gkae1105.
Chomiak AA, Tiedemann RL, Liu Y, Kong X, Cui Y, Wiseman AK, Thurlow KE, Cornett EM, Topper MJ, Baylin SB, Rothbart SB. 2024. Select EZH2 inhibitors enhance viral mimicry effects of DNMT inhibition through a mechanism involving NFAT:AP-1 signaling. Sci Adv 10(13).
2023
Luda KM, Longo J, Kitchen-Goosen SM, Duimstra LR, Ma EH, Watson MJ, Oswald BM, Fu Z, Madaj Z, Kupai A, Dickson BM, DeCamp LM, Dahabieh MS, Compton SE, Teis R, Kaymak I, Lau KH, Kelly DP, Puchalska P, Williams KS, Krawczyk CM, Lévesque D, Boisvert FM, Sheldon RD, Rothbart SB, Crawford PA, Jones RG. 2023. Ketolysis drives CD8+ T cell effector function through effects on histone acetylation. Immunity.
Compton SE, Kitchen-Goosen SM, DeCamp LM, Lau KH, Mabvakure, Vos M, Williams KS, Wong KK, Shi X, Rothbart SB, Krawczyk CK, Jones RG. 2023. LKB1 controls inflammatory potential through CRTC2-dependent histone acetylation. Mol Cell.
Berryhill CA, Hanquier JN, Doud EH, Cordeiro-Spinetti E, Dickson BM, Rothbart SB, Mosley AL, Cornett EM. 2023. Global lysine methylome profiling using systematically characterized affinity reagents. Sci Rep13(1):377.
2022
Reske JJ, Wilson MR, Armistead B, Harkins S, Peres C, Hrit J, Adams M, Rothbart SB, Missmer SA, Fazleabas AT, Chandler RL. 2022. ARID1A-dependent maintenance of H3.3 is required for repressive CHD4-ZMYND8 chromatin interactions at super-enhancers. BMC Biol 20(1):209.
2021
Vaughan RM, Kupai A, Rothbart SB. 2021. Chromatin regulation through ubiquitin and ubiquitin-like histone modifications. Trends Biochem Sci 46(4): 258–269.
Enríquez P, Krajewski K, Strahl BD, Rothbart SB, Dowen R, Rose RB. 2021. Binding specificity and function of the SWI/SNF subunit SMARCA4 bromodomain interaction with acetylated histone H3K14. J Biol Chem 101145.
Slaughter MJ, Shanle EK, Khan A, Chua KF, Hong T, Boxer LD, Allis CD, Josefowicz SZ, Garcia BA, Rothbart SB, Strahl BD, Davis IJ. 2021. HDAC inhibition results in widespread alteration of the histone acetylation landscape and BRD4 targeting to gene bodies. Cell Rep 34(3):108638.
2020
Vaughan RM, Kupai A, Rothbart SB. 2020. Chromatin regulation through ubiquitin and ubiquitin-like histone modifications. Trends Biochem Sci.
Franks JL, Martinez-Chacin RC, Wang X, Tiedemann RL, Bonacci T, Choudhury R, Bolhuis DL, Enrico TP, Mouery RD, Damrauer JS, Yan F, Harrison JS, Major MB, Hoadley KA, Suzuki A, Rothbart SB, Brown NG, Emanuele MJ. 2020. In silico APC/C substrate discovery reveals cell cycle-dependent degradation of UHRF1 and other chromatin regulators. PLoS Biol.
Fan H, Atiya HI, Wang Y, Pisanic TR, Wang TH, Shih IM, Foy KK, Frisbie L, Buckanovich RJ, Chomiak AA, Tiedemann RL, Rothbart SB, Chandler C, Shen H, Coffman LG. 2020. Epigenomic reprogramming toward mesenchymal-epithelial transition in ovarian-cancer-associated mesenchymal stem cells drives metastasis. Cell Rep.
Vaughan RM, Kupai A, Foley CA, Sagum CA, Tibben BM, Eden HE, Tiedemann RL, Berryhill CA, Patel V, Shaw KM, Krajewski K, Strahl BD, Bedford MT, Frye SV, Dickson BM, Rothbart SB. 2020. The histone and non-histone methyllysine reader activities of the UHRF1 tandem Tudor domain are dispensable for the propagation of aberrant DNA methylation patterning in cancer cells. Epigenetics Chromatin 13(1):44.
Armache A, Yang S, Martínez de Paz A, Robbins LE, Durmaz C, Cheong JQ, Ravishankar A, Daman AW, Ahimovic DJ, Klevorn T, Yue Y, Arslan T, Lin S, Panchenko T, Hrit J, Wang M, Thudium S, Garcia BA, Korb E, Armache KJ, Rothbart SB, Hake SB, Allis CD, Li H, Josefowicz SZ. 2020. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 583(7818):852–857.
Rothbart SB, Baylin SB. 2020. Epigenetic therapy for epithelioid sarcoma. Cell 181(2):211.
Dickson BM, Tiedemann RL, Chomiak AA, Cornett EM, Vaughan RM, Rothbart SB. 2020. A physical basis for quantitative ChIP-sequencing. J Biol Chem 295(47):15826–15837.
*Selected as an Editor’s Pick and featured on the cover
Kupai A, Vaughan RM, Dickson BM, Rothbart SB. 2020. A degenerate peptide library approach to reveal sequence determinants of methyllysine-driven protein interactions. Front Cell Dev Biol.
2019
Colino-Sanguino Y, Cornett EM, Moulder D, Smith GC, Hrit J, Cordeiro-Spinetti E, Vaughan RM, Krajewski K, Rothbart SB*, Clark SJ*, Valdés-Mora F*. 2019. A read/write mechanism connects p300 Bromodomain function to H2A.Z acetylation. iScience S2589-0042(19)30434-1.
*Co-corresponding authors
Cornett E, Ferry L, Defossez PA, Rothbart SB. 2019. Lysine methylation regulators moonlighting outside the epigenome. Mol Cell.
Vaughan RM, Rothbart SB*, Dickson BM*. 2019. The finger loop of the SRA domain in the E3 ligase UHRF1 is a regulator of ubiquitin targeting and is required for maintaining DNA methylation. J Biol Chem.
*Co-corresponding authors
Zhang Y, Jang Y, Lee JE, Ahn J, Xu L, Holden MR, Cornett EM, Krajewski K, Klein BJ, Wang SP, Dou Y, Roeder RG, Strahl BD, Rothbart SB, Shi X, Ge K, Kutateladze TG. 2019. Selective binding of the PHD6 finger of MLL4 to histone H4K16ac links MLL4 and MOF. Nat Commun 10(1):2314.
Kong X*, Chen J*, Xie W*, Brown SM, Cai Y, Wu K, Fan D, Nie Y, Yegnasubramanian S, Tiedemann RL, Tao Y, Yen RWC, Topper MJ, Zahnow CA, Easwaran H, Rothbart SB#, Xia L#, Baylin SB#. 2019. Defining UHRF1 domains that support maintenance of human colon cancer DNA methylation and oncogenic properties. Cancer Cell.
*These authors contributed equally
#Co-corresponding authors
Selected as a paper of the month by National Institute of Environmental Health Sciences
Harlow ML*, Chassé MH*, Boguslawski EA, Sorensen KM, Gedminas MJ, Goosen JM, Rothbart SB, Taslim C, Lessnick SL, Peck A, Madaj ZB, Bowman MJ, Grohar PJ. 2019. Trabectedin inhibits EWS-FLI1 and evicts SWI/SNF from chromatin in a schedule-dependent manner. Clin Cancer Res.
*Equal contributions
2018
Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, Li X, Chen W, Xue Y, Zhong Z, Yen L, Barshop WD, Rayatpisheh S, Gallego-Bartolome J, Groth M, Wang Z, Wohlschlegel JA, Du J, Rothbart SB, Butter F, Jacobsen SE. 2018. A DNA methylation reader complex that enhances gene transcription. Science 362(6419):1182–1186.
Cornett EM, Dickson BM, Krajewski K, Spellmon N, Umstead A, Vaughan RM, Shaw KM, Versluis PP, Cowles MW, Brunzelle J, Yang Z, Vega IE, Sun ZW, Rothbart SB. 2018. A functional proteomics platform to reveal the sequence determinants of lysine methyltransferase substrate selectivity. Sci Adv.
Shah RN, Grzybowski AT, Cornett EM, Johnstone AL, Dickson BM, Boone BA, Cheek MA, Cowles MW, Maryanski D, Meiners MJ, Tiedemann RL, Vaughan RM, Arora N, Sun ZW, Rothbart SB*, Keogh MC*, Ruthenberg AJ*. 2018. Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol Cell.
Video abstract
*Equal contributions
Javasky E, Shamir I, Gandhi S, Egri S, Sandler O, Rothbart SB, Kaplan N, Jaffe JD, Goren A, Simon I. 2018. Study of mitotic chromatin supports a model of bookmarking by histone modifications and reveals nucleosome deposition patterns. Genome Res.
Vaughan RM, Dickson BM, Whelihan MF, Johnstone AL, Cornett EM, Cheek MA, Ausherman CA, Cowles MW, Sun ZW, Rothbart SB. 2018. Chromatin structure and its chemical modifications regulate the ubiquitin ligase substrate selectivity of UHRF1. Proc Natl Acad Sci U S A.
Vaughan RM, Dickson BM, Cornett EM, Harrison JS, Kuhlman B, Rothbart SB. 2018. Comparative biochemical analysis of UHRF proteins reveals molecular mechanisms that uncouple UHRF2 from DNA methylation maintenance. Nuc Acids Res.
Zhang ZM, Lu R, Wang P, Yu Y, Chen D, Gao L, Liu S, Ji D, Rothbart SB, Wang Y, Wang GG, Song J. 2018. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature.
2017
Veland N, Hardikar S, Zhong Y, Gayatri S, Dan J, Strahl BD, Rothbart SB, Bedford MT, Chen T. 2017. The arginine methyltransferase PRMT6 regulates DNA methylation and contributes to global DNA hypomethylation in cancer. Cell Rep 21(2):3390–3397.
Cornett EM, Dickson BM, Rothbart SB. 2017. Analysis of histone antibody specificity with peptide microarrays. JoVE (126):doi: 10.3791/55912.
Ma H, Duan J, Ke J, He Y, Gu X, Xu TH, Yu H, Wang Y, Brunzelle JS, Jiang Y, Rothbart SB, Xu HE, Li J, Melcher K. 2017. A D53 repression motif induces oligomerization of TOPLESS copressors and promotes assembly of a corepressor-nucleosome complex. Sci Adv.
Shanle EK, Shinsky SA, Bridgers JB, Bae N, Sagum C, Krajewski K, Rothbart SB, Bedford MT, Strahl BD. 2017. Histone peptide microarray screen of chromo and Tudor domains defines new histone lysine methylation interactions. Epigenetics Chromatin 10:12.
2016
Busby M, Xue C, Li C, Farjourn Y, Gienger E, Yofe I, Gladden A, Epstein CB, Cornett EM, Rothbart SB, Nusbaum C, Goren A. 2016. Systematic comparison of monoclonal versus polyclonal antibodies for mapping histone modifications by ChIP-seq. Epigenomics Chromatin 9:49.
Dickson BM, de Waal PW, Ramjan ZH, Xu HE, Rothbart SB. 2016. A fast, open source implementation of adaptive biasing potential uncovers a ligand design strategy from the chromatin regulator BRD4. J Chem Phys 145:154113.
Strikoudis A, Lazaris C, Trimarchi T, Galvoa Neto AL, Yang Y, Ntziachristos P, Rothbart SB, Buckley S, Dolgalev I, Stadtfeld M, Strahl BD, Dynlacht BD, Tsirigos A, Iannia Aifantis. 2016. Regulation of transcriptional elongation in pluripotency and cell differentiation by the PHD-finger protein Phf5a. Nat Cell Biol 18(11):1127–1138.
Andrews FH, Tong Q, Sullivan KD, Cornett EM, Zhang Y, Ali M, Ahn J, Pandey A, Guo AH, Strahl BD, Costello JC, Espinosa JM, Rothbart SB, Kutateladze. 2016. Multivalent chromatin engagement and inter-domain crosstalk regulate MORC3 ATPase. Cell Rep 16(12):3195–3207.
Harrison J, Cornett E, Goldfarb D, DaRosa P, Li Z, Yan F, Dickson B, Guo A, Cantu D, Kaustov L, Brown P, Arrowsmith C, Klevit R, Erie D, Major B, Krajewski K, Kulhman B, Strahl B, Rothbart SB. 2016. Hemi-methylated DNA regulates DN methylation inheritance through allosteric activation of H3 ubiquitylation for UHRF1. eLife.
Cornett EM, Dickson BM, Vaughan RM, Krishnan S, Trievel RC, Strahl BD, Rothbart SB. 2016. Substrate specificity profiling of histone-modifying enzymes by peptide microarray. Methods Enzymol 574:31–52.
Dickson BM, Cornett EM, Ramjan Z, Rothbart SB. 2016. ArrayNinja: An open source platform for unified planning and analysis of microarray experiments. Methods Enzymol 574:53–57.
2015
Agarwal S, Bell C, Rothbart SB, Moran RG. 2015. AMPK control of mTORC1 is p53- and TSC2-independent in pemetrexed-treated carcinoma cells. J Biol Chem 290(46):27473–27486.
Cliffe AR, Arbuckle JH, Vogel JL, Geden MJ, Rothbart SB, Cusack CL, Kristie TM, Deshmukh M. 2015. Neuronal stress pathway mediating a histone methyl/phospho switch is required for herpes simplex virus reactivation. Cell Host Microbe. 18(6):649–658.
Zhang ZM, Rothbart SB, Allison DF, Cai Q, Harrison JS, Li L, Wang Y, Strahl BD, Wang GG, Song J. 2015. An allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1. Cell Rep 12(9):1400–1406.
Simon JM, Parker JS, Liu F, Rothbart SB, Ait-Si-Ali S, Strahl BD, Jin J, Davis IJ, Mosley AL, Pattenden SG. 2015. A role for widely interspaced zinc finger (WIZ) in retention of the G9a methyltransferase on chromatin. J Biol Chem. 290(43):26088–26102.
Ali M, Daze KD, Strongin DE, Rothbart SB, Rincon-Arano H, Allen HF, Li J, Strahl BD, Hof F, Kutatladze TG. 2015. Molecular insight into inhibition of the methylated histone-plant homeodomain complexes by calixarenes. J Biol Chem. 10(4):1072–1081.
Rothbart SB*, Dickson BM, Raab JR, Grzybowski AT, Krajewski K, Guo AH, Shanle EK, Josefowicz SZ, Fuchs SM, Allis CD, Magnuson TR, Ruthenburg AJ, Strahl BD*. 2015. An interactive database for the assessment of histone antibody specificity. Mol Cell. 59(3):502–511
*Equal contributions
Perfetti MT, Baughman BM, Dickson BM, Mu Y, Cui G, Mader P, Dong A, Norris JL, Rothbart SB, Strahl BD, Brown PJ, Janzen WP, Arrowsmith CH, Mer G, McBride KM, James LI, Frye SV. 2015. Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1. ACS Chem Biol 10(4):1072—1081
Rothbart SB, Dickson BM, and Strahl BD. 2015. From histones to ribosomes: A chromatin regulator tangoes with translation. Cancer Discov 5(3):228–230
Tong Q, Mazur SJ, Rincon-Arano H, Rothbart SB, Kuznetsov DM, Cui G, Liu WH, Gete Y, Klein BJ, Jenkins L, Mer G, Kutateladze AG, Strahl BD, Groudine M, Appella E, Kutateladze TG. 2015. An acetyl-methyl switch drives a conformational change in p53. Structure. 23(2):322-331
Tong Q, Cui G, Botuyan MV, Rothbart SB, Hayashi R, Musselman CA, Singh N, Appella E, Strahl BD, Mer G, Kutateladze TG. 2015. Structural plasticity of methyllysine recognition by the tandem Tudor domain of 53BP1. Structure. 23(2):312-322
ArrayNinja is a portable, open source and interactive application that unifies the planning, visualization, and analysis of custom microarray experiments.
ArrayNinja is distributed as a virtual machine (VM) that runs a local and private web server. Because the tools are packaged as a VM and utilize a web browser as a user interface, ArrayNinja is platform agnostic.
Download and installation
Virtual Machine
The ArrayNinja VM is built to use VirtualBox, which is a free tool for building and running virtual machines. VirtualBox must be installed before the ArrayNinja VM can be used.
To download the ArrayNinja VM, visit https://hub.docker.com/r/bradleydickson/arrayninja/.
To run ArrayNinja, unzip the VM and load the VM into virtualbox. Once the VM is running, navigate your web browser to the localhost:2080.
Docker Application
ArrayNinja can be run on linux platforms via a docker application. The docker can be found at Docker Hub and it can be installed by pulling bradleydickson/arrayninja.
The docker app should be started with command:
docker run -d -p 2080:80 bradleydickson/arrayninja
License
Academic and not-for-profit
ArrayNinja is released under GNU General Public License, version 2. This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; version 2 of the License.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA 02110-1301, USA.
Contact
For business and licensing inquiries, please contact [email protected].
For scientific inquiries, please contact [email protected].
Citations
Dickson BM, Cornett EM, Ramjan Z, Rothbart SB. 2016. ArrayNinja: An open source platform for unified planning and analysis of microarray experiments. Methods Enzymol 574:53–57.
Publications
Dickson BM, Cornett EM, Ramjan Z, Rothbart SB. 2016. ArrayNinja: An open source platform for unified planning and analysis of microarray experiments. Methods Enzymol 574:53–57.
Cornett EM, Dickson BM, Vaughan RM, Krishnan S, Trievel RC, Strahl BD, Rothbart SB. 2016. Substrate specificity profiling of histone-modifying enzymes by peptide microarray. Methods Enzymol 574:31–52.
fABMACS is a flexible adaptively biased flavor of GROMACSv5.0.5. fABMACS was developed to maximize scalability of adaptive biasing so that better efficiency can be achieved when simulation systems are distributed across a large computing infrastructure.
Download and installation
fABMACS is available for download and use. Instructions and example inputs (for the ligand-protein system in the publication) are available as well. Please visit https://github.com/BradleyDickson/fABMACS.
License
Academic and not-for-profit
fABMACS is released under GNU General Public License, version 2. This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; version 2 of the License.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA 02110-1301, USA.
Contact
For questions about fABMACS or commercial use, contact [email protected].
Citations
Dickson BM, de Waal PW, Ramjan ZH, Xu HE, Rothbart SB. In press. A fast, open source implementation of adaptive biasing potential uncovers a ligand design strategy from the chromatin regulator BRD4. J Chem Phys. Article
Access to high-quality antibodies is a necessity for the study of histones and their posttranslational modifications (PTMs). The Histone Antibody Specificity Database is an online and expanding resource that catalogs the behavior of widely used, commercially available histone antibodies by peptide microarray. This interactive web portal provides a critical resource to the biological research community that routinely uses these antibodies as detection reagents for a wide range of applications.
Contact
Citations
Rothbart SB, Dickson BM, Raab JR, Grzybowski AT, Krajewski K, Guo AH, Shanle EK, Josefowicz SZ, Fuchs SM, Allis CD, Magnuson TR, Ruthenberg AJ, Strahl BD. 2015. An interactive database for the assessment of histone antibody specificity. Molecular Cell 59(3):502–511.
Publications
Dickson BM, Cornett EM, Ramjan Z, Rothbart SB. In press. ArrayNinja: An open source platform for unified planning and analysis of microarray experiments. Methods Enzymol.
Cornett EM, Dickson BM, Vaughan RM, Krishnan S, Trievel RC, Strahl BD, Rothbart SB. In press. Substrate specificity profiling of histone-modifying enzymes by peptide microarray. Methods Enzymol.
Rothbart SB, Dickson BM, Raab JR, Grzybowski AT, Krajewski K, Guo AH, Shanle EK, Josefowicz SZ, Fuchs SM, Allis CD, Magnuson TR, Ruthenberg AJ, Strahl BD. 2015. An interactive database for the assessment of histone antibody specificity. Molecular Cell 59(3):502–511.

We are continually seeking highly motivated individuals with a strong publication record to join us in our energetic, interactive and fast-paced environment. A Ph.D. with molecular biology experience is required to apply. Additional experience in biochemistry, proteomics, microarray and/or cell biology is preferred (but not necessary) as we seek individuals with high training potential. Interested candidates should email Dr. Rothbart to inquire. Graduate students interested in joining the lab are encouraged to apply through Van Andel Institute Graduate School.


Bradley Dickson, Ph.D.
VAI Staff Scientist

Joel Hrit, Ph.D.
Research Scientist, Department of Epigenetics
Regulatory mechanisms of maintenance DNA methylation by DNMT1

Alison Lanctot Chomiak, Ph.D.
Research Scientist, Department of Epigenetics


Yanqing Liu, M.Sc.
Ph.D. Student, VAI Graduate School
Research Focus: Mechanisms and therapeutic vulnerabilities of DNMTi-associated epigenetic plasticity

Amy Nuffesse
Senior Administrative Assistant II, Department of Epigenetics

Tian (Devin) Qiu, M.S.
Ph.D. Student, VAI Graduate School
Research Focus: Spatial multi-omics profiling of DNMT inhibitor-treated solid tumor biopsies
Vincent Sartori
Ph.D. Student, VAI Graduate School
Research focus to be determined.

Kate Thurlow, M.Sc.
Ph.D. Student, VAI Graduate School
Thesis project title to be determined

Rochelle Tiedemann, Ph.D.
Research Scientist, Department of Epigenetics

Ashley Wiseman, M.S.
Laboratory Manager, Department of Epigenetics

 Chromatin and Epigenetic Regulation
Chromatin and Epigenetic Regulation
